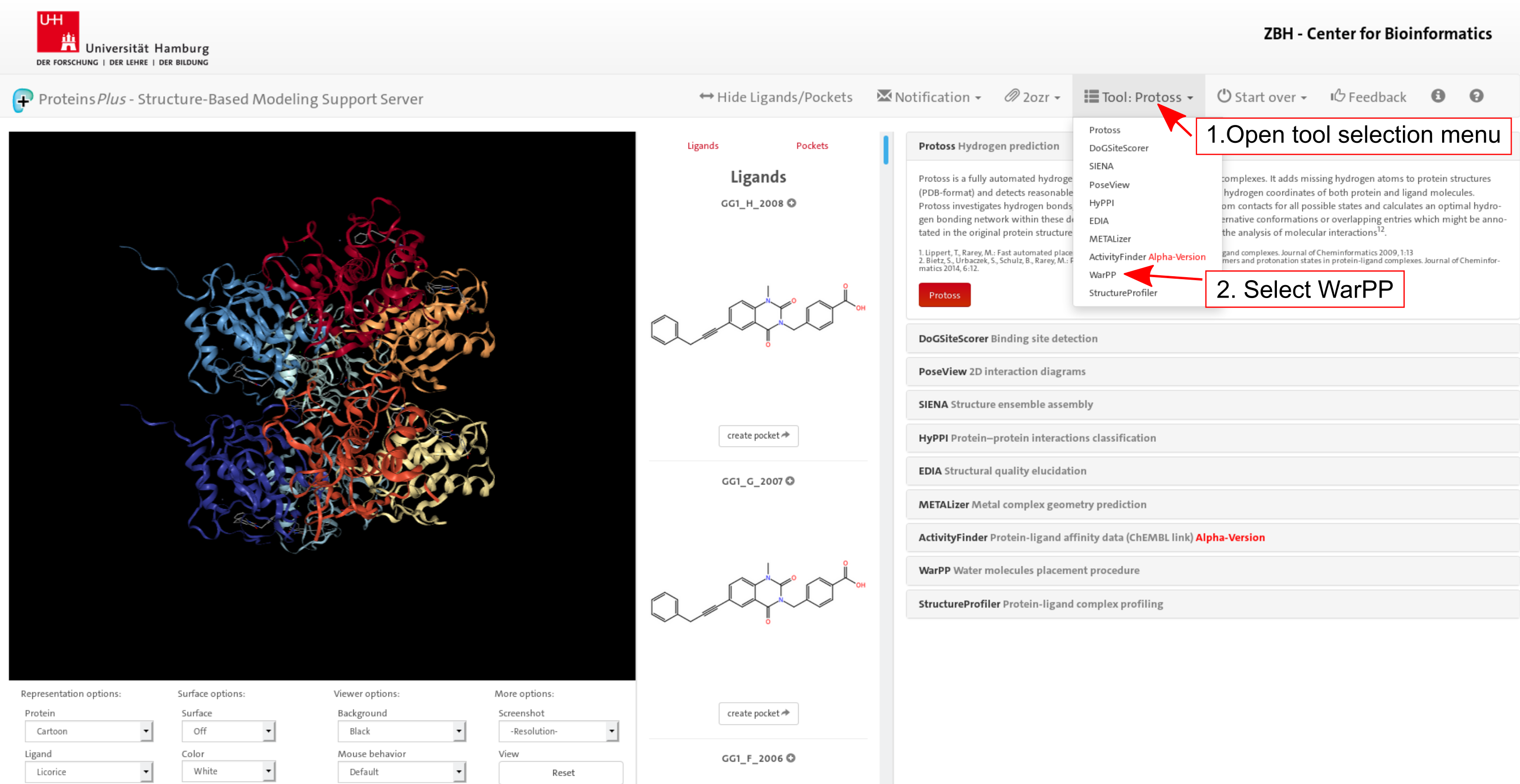
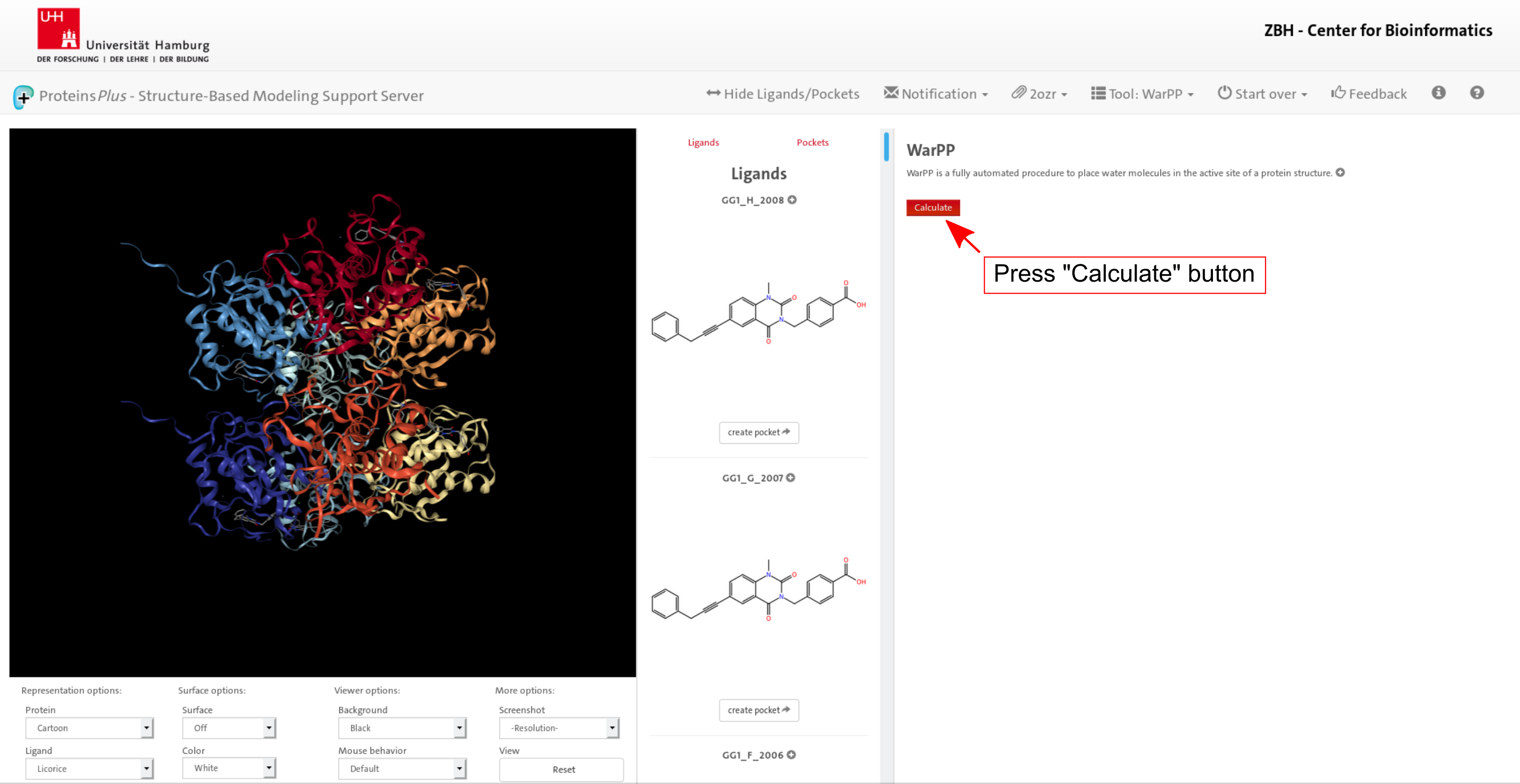
1. Target definition
All tools in the ProteinsPlus server work on a structure of a protein or a nucleic acid.
In the first step, you need to define this structure either by selecting a structure form the
Protein Data Bank (PDB) via its PDB code (e.g. 1xm6) or upload any other structure in PDB format.
Additional ligand molecules can be provided in SDF format.
Click on image to jump to that page.
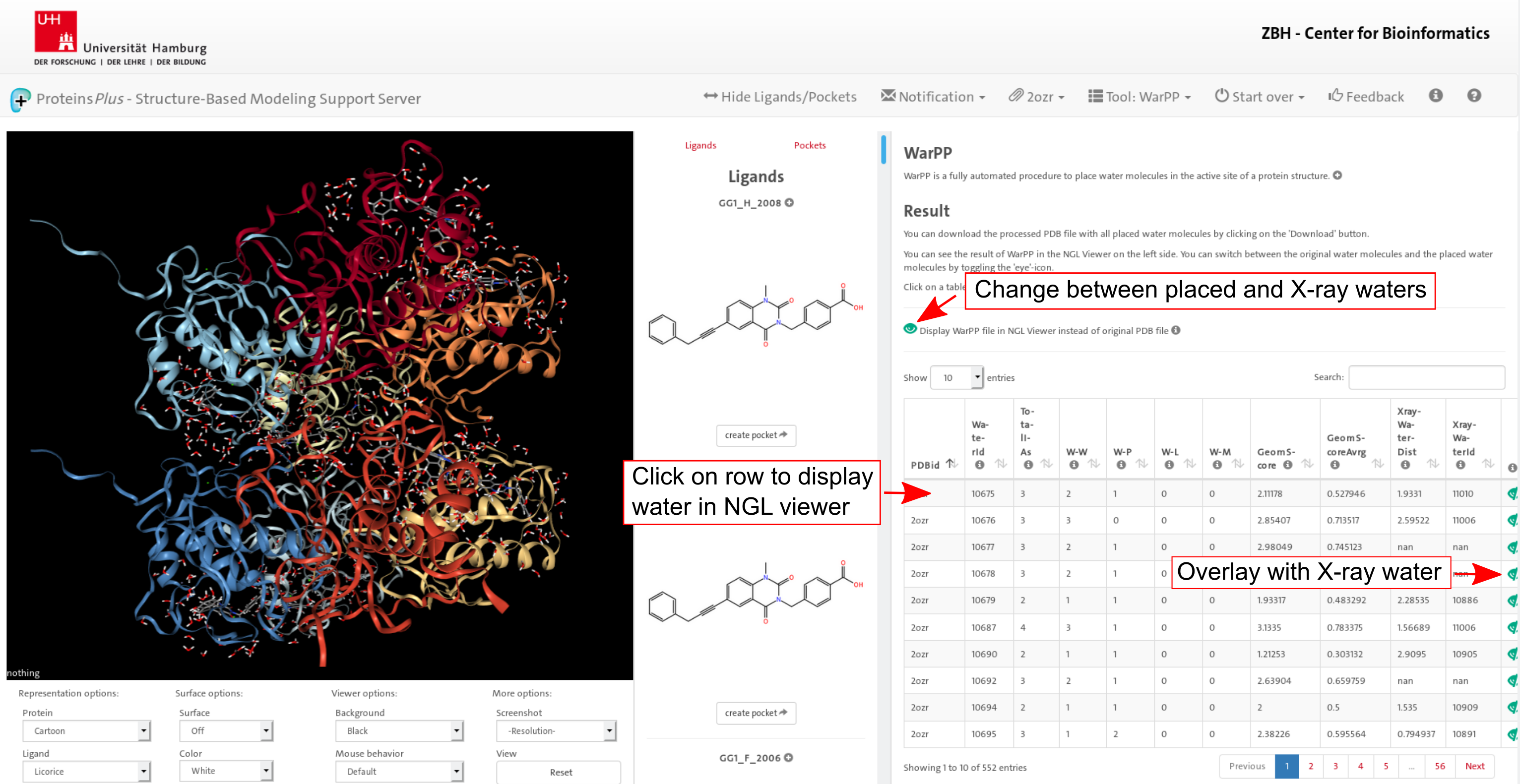
4. Result
When the WarPP calculation is finished, the placed water molecules are displayed
in the NGL viewer. The original PDB file can be displayed by toggling the eye icon. Additionally several
characteristics about each water molecule are tabulated - the number of interactions based on the NAOMI
library, the type of interaction partners (protein, ligand, metal, or water) as well as the closest X-ray
water. For each placed water molecule the original X-ray water can be displayed by toggling the eye icon at
the end of each row. All information can be additionally downloaded using the download button.
Click on image to jump to that page.
Publication
For additional information on WarPP, please refer to the corresponding publication:
Nittinger, E.; Flachsenberg, F.; Bietz, S.; Lange, G.; Klein, R.; Rarey, M.(2018).
Placement of Water Molecules in Protein Structures: From Large-Scale Evalutations to Single-Case Examples.
Journal of Chemical Information and Modeling, 58 (8), pp 1625-1637.